26+ protein charge calculator
Web The Protein Charge Calculator calculates the charge of a protein using the Ohshima equation for monovalent salts. Web The calculator is also useful for monitoring protein intake for those with kidney disease liver disease diabetes or other conditions in which protein intake is a factor.
De Novo Amino Acid Sequence Elucidation Of Protein G E By Combined Top Down And Bottom Up Mass Spectrometry Springerlink
If plotResults FALSE a.

. Web The calculators are a series of tools in the Zetasizer software that enable the user to calculate a number of extra parameters such as protein charge and the DLS interaction. Web Draw a protein charge plot read the manual Unshaded fields are optional and can safely be ignored. Web Peptide Analyzing Tool.
Web The environmental pH used to calculate residue charge. Web Protein calculator N-terminus Sequence C-terminus AA code used Disulphide connectivity Show abbreviations 20 standard amino acids modified amino acids unusual amino acids. Web Calculate masses of b and y daughter ions.
The tool can auto-detect most formats of single-letter and three-letter amino acid sequences. Use Monoisotopic Masses Not Isotopic Averages Methionine to Selenomethionine. Once this is done calculating the net charge is straightforward.
Anyone has any idea how to calculate protein surface charges electrostatic. Input section Select an input sequence. Web Complete Protein Calculator - A tool to make sure you are getting all the essential amino acids.
Web Compute pIMw for Swiss-ProtTrEMBL entries or a user-entered sequence Please enter one or more UniProtKBSwiss-Prot protein identifiers ID eg. Type or paste the sequence of a peptide and click Analyze. Perhaps any online tools to do that.
Web ProtParam References Documentation is a tool which allows the computation of various physical and chemical parameters for a given protein stored in Swiss-Prot or TrEMBL or. Example Recipes Firm Tofu Rice and Broccoli Apples Bananas and Almonds. Web How to calculate protein surface charge.
Logical value FALSE by default. Web It is your job to calculate the proportion of each amino acid that is ionized at a given pH. At pH 3 K R H are and DE have no charge so add up all of the KRH in the sequence and that is your net charge at pH 3 At pH 6 K R.
This determines what is returned. Web The NIST Mass and Fragment Calculator is a program written in Visual Basic which calculates the mass of an input peptide or protein sequence along with mz ions. It takes f κa from the Henry equation into.
Use one of the following three fields. Web To calculate the charge at different pH. Web Isoelectric Point Calculator 20 IPC 20 - is a server for the prediction of isoelectric points and pKa values using a mixture of deep learning and support vector regression models.

Calculating Net Charge On Proteins Youtube
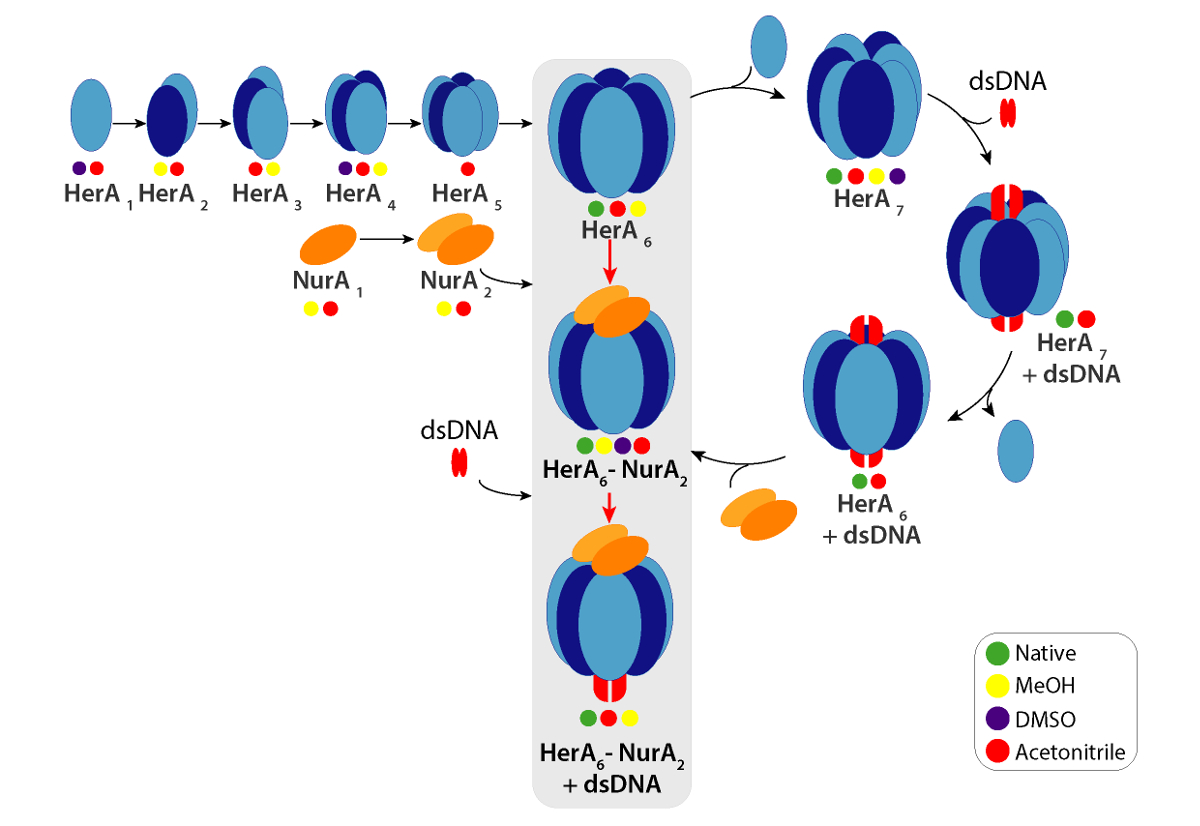
Analyzing Protein Architectures And Protein Ligand Complexes By Integrative Structural Mass Spectrometry Video
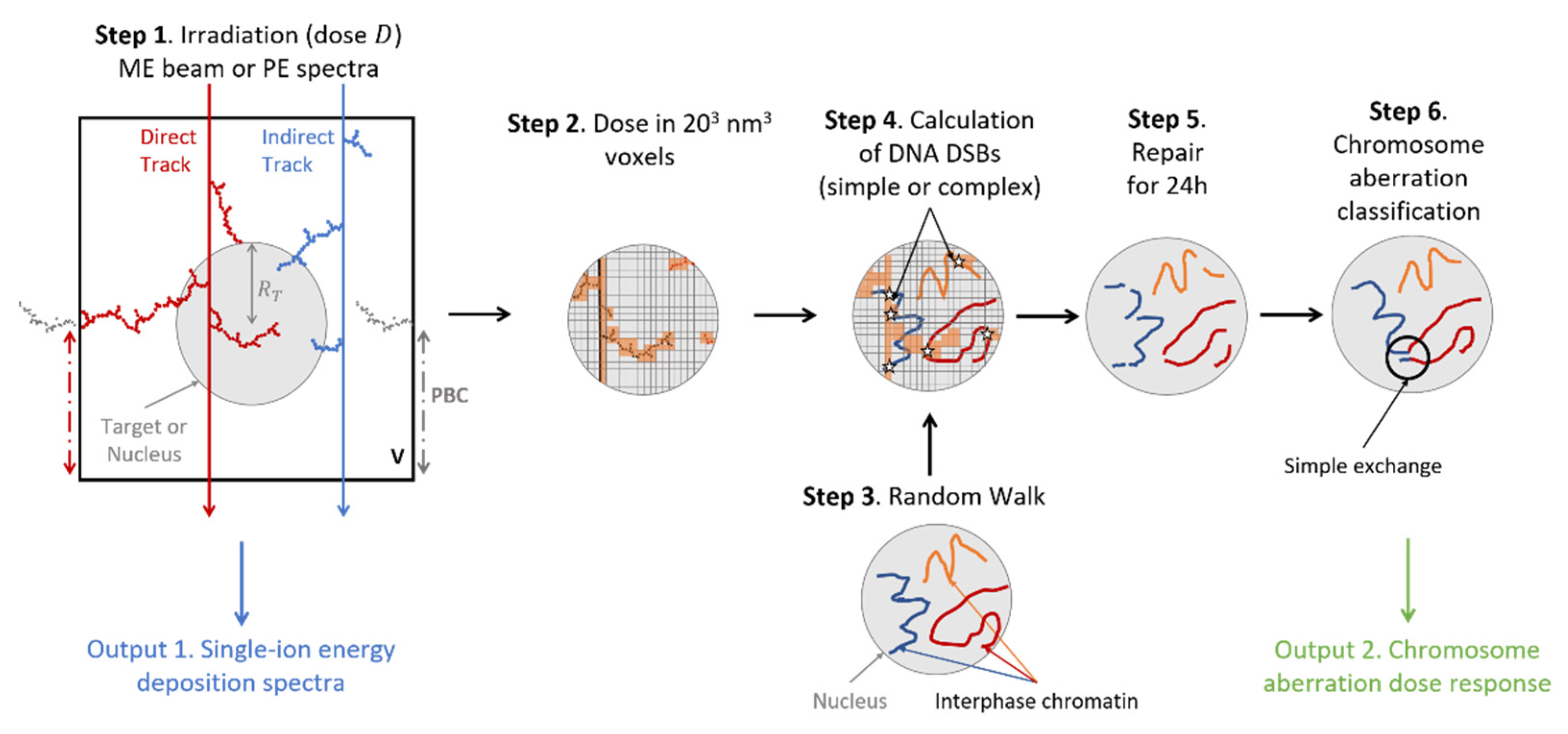
Life Free Full Text Impact Of Radiation Quality On Microdosimetry And Chromosome Aberrations For High Energy Gt 250 Mev N Ions
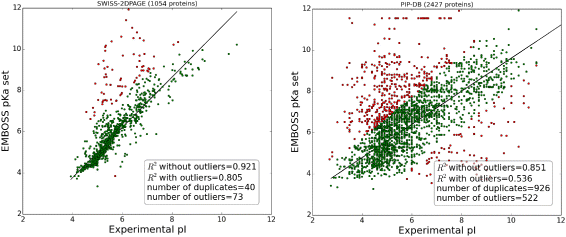
Ipc Isoelectric Point Calculator Biology Direct Full Text
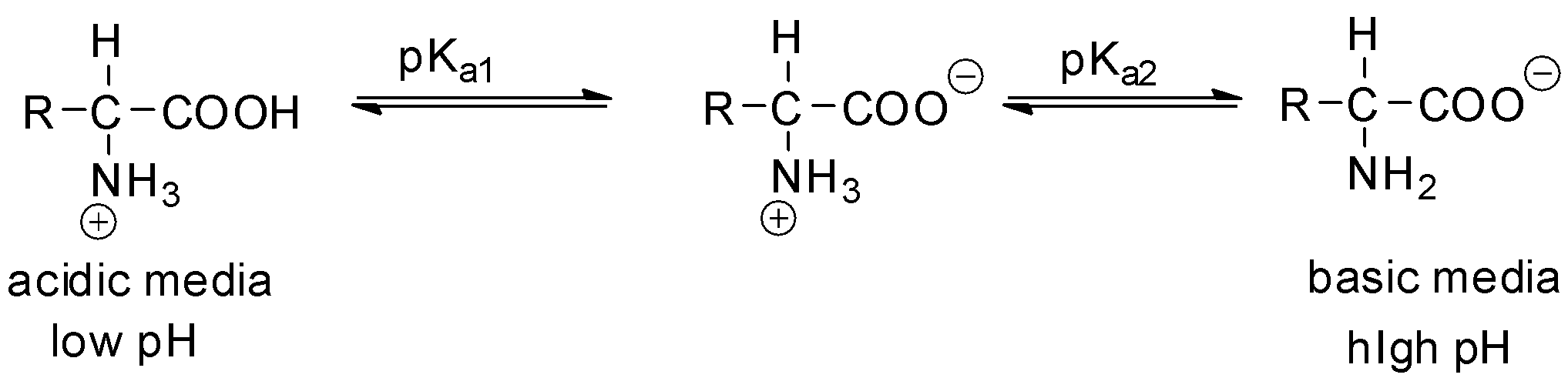
Find The Isoelectric Point Pi Of Lysine A 5 56 B 9 74 C 6 25 D 0

Native Mass Spectrometry Reveals The Conformational Diversity Of The Uvr8 Photoreceptor Pnas
Ipc Isoelectric Point Calculation Of Proteins And Peptides

Native Mass Spectrometry Reveals The Conformational Diversity Of The Uvr8 Photoreceptor Pnas

How To Calculate Peptide Charge And Isoelectric Point Mcat Trick Youtube

Calculating Net Protein Charge The Problem A Protein S Net Charge Depends On The Number Of Charged Amino Acids It Contains And The Ph Of Its Environment Ppt Download
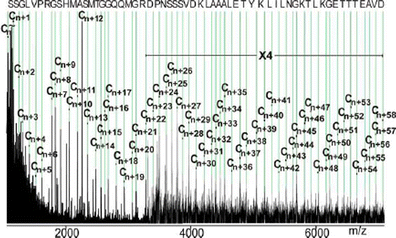
De Novo Amino Acid Sequence Elucidation Of Protein G E By Combined Top Down And Bottom Up Mass Spectrometry Springerlink
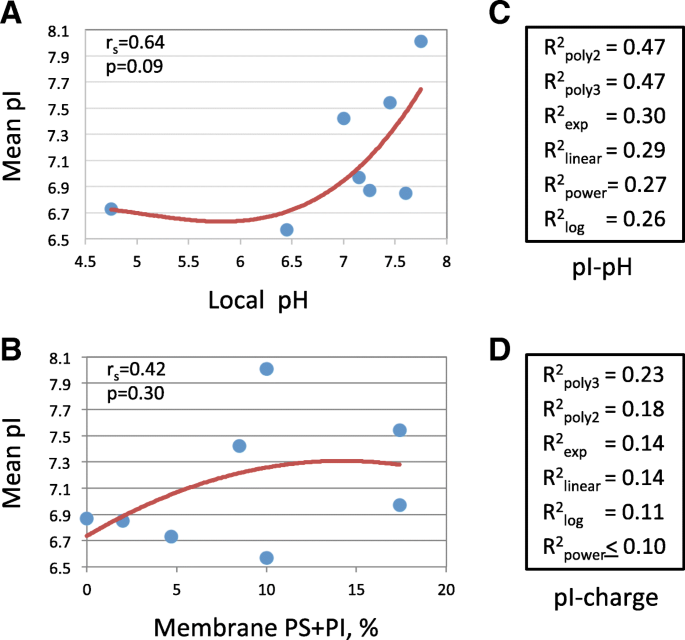
Localization Specific Distributions Of Protein Pi In Human Proteome Are Governed By Local Ph And Membrane Charge Bmc Molecular And Cell Biology Full Text
Chem 440 Lecture 6
Peptide Calculator

Isotopic Resolution Of Protein Complexes Up To 466 Kda Using Individual Ion Mass Spectrometry Analytical Chemistry
Peptide Calculator

Protein Derivatization And Sequential Ion Ion Reactions To Enhance Sequence Coverage Produced By Electron Transfer Dissociation Mass Spectrometry Sciencedirect